Abstract
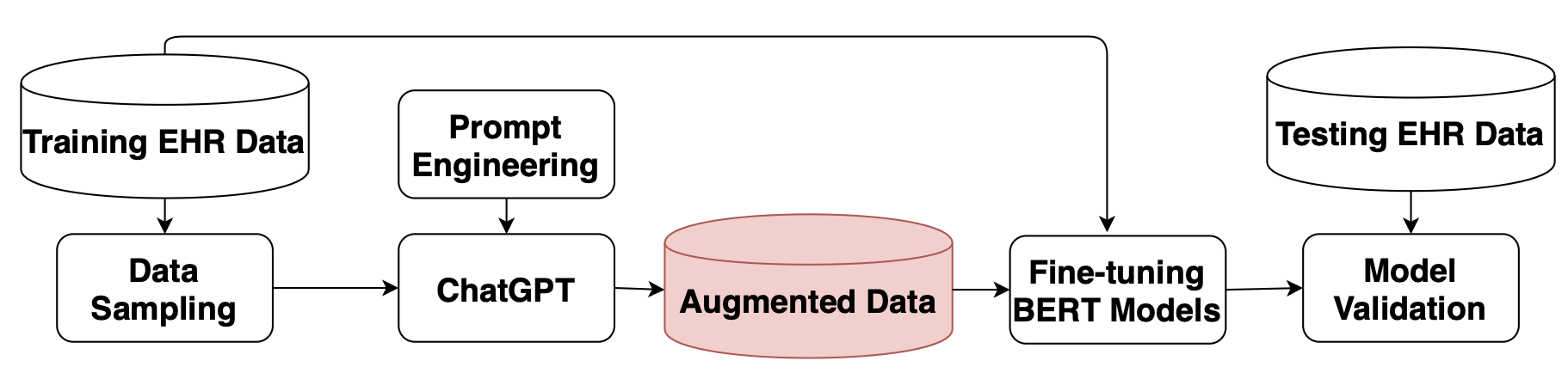
The identification of key factors such as medications, diseases, and relationships within electronic health records and clinical notes has a wide range of applications in the clinical field. In the N2C2 2022 competitions, various tasks were presented to promote the identification of key factors in electronic health records (EHRs) using the Contextualized Medication Event Dataset (CMED). Pretrained large language models (LLMs) demonstrated exceptional performance in these tasks. This study aims to explore the utilization of LLMs, specifically ChatGPT, for data augmentation to overcome the limited availability of annotated data for identifying the key factors in EHRs. Additionally, different pre-trained BERT models, initially trained on extensive datasets like Wikipedia and MIMIC, were employed to develop models for identifying these key variables in EHRs through fine-tuning on augmented datasets. The experimental results of two EHR analysis tasks, namely medication identification and medication event classification, indicate that data augmentation based on ChatGPT proves beneficial in improving performance for both medication identification and medication event classification.