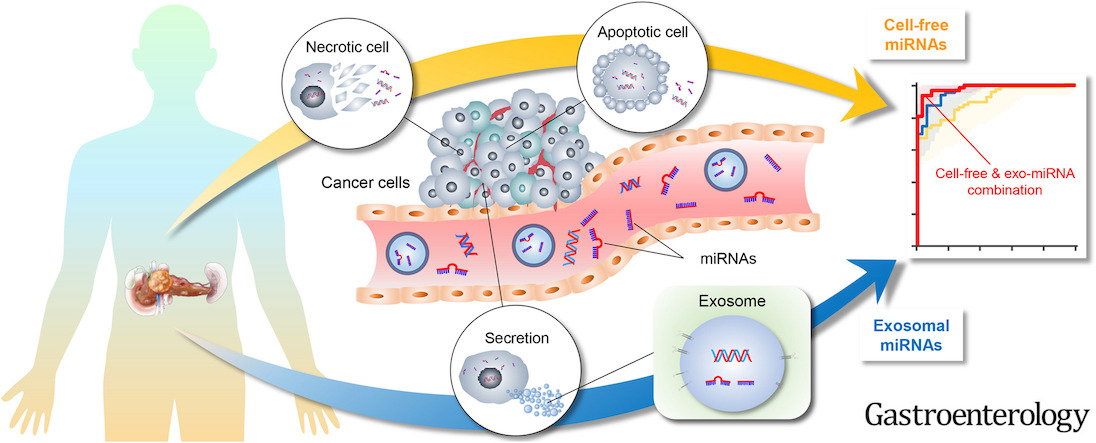
Abstract
Background & aims: Pancreatic ductal adenocarcinoma (PDAC) incidence is rising worldwide, and most patients present with an unresectable disease at initial diagnosis. Measurement of carbohydrate antigen 19-9 (CA19-9) levels lacks adequate sensitivity and specificity for early detection; hence, there is an unmet need to develop alternate molecular diagnostic biomarkers for PDAC. Emerging evidence suggests that tumor-derived exosomal cargo, particularly micro RNAs (miRNAs), offer an attractive platform for the development of cancer-specific biomarkers. Herein, genomewide profiling in blood specimens was performed to develop an exosome-based transcriptomic signature for noninvasive and early detection of PDAC.
Methods: Small RNA sequencing was undertaken in a cohort of 44 patients with an early-stage PDAC and 57 nondisease controls. Using machine-learning algorithms, a panel of cell-free (cf) and exosomal (exo) miRNAs were prioritized that discriminated patients with PDAC from control subjects. Subsequently, the performance of the biomarkers was trained and validated in independent cohorts (n = 191) using quantitative real-time polymerase chain reaction (qRT-PCR) assays.
Results: The sequencing analysis initially identified a panel of 30 overexpressed miRNAs in PDAC. Subsequently using qRT-PCR assays, the panel was reduced to 13 markers (5 cf- and 8 exo-miRNAs), which successfully identified patients with all stages of PDAC (area under the curve [AUC] = 0.98 training cohort; AUC = 0.93 validation cohort); but more importantly, was equally robust for the identification of early-stage PDAC (stages I and II; AUC = 0.93). Furthermore, this transcriptomic signature successfully identified CA19-9 negative cases (<37 U/mL; AUC = 0.96), when analyzed in combination with CA19-9 levels, significantly improved the overall diagnostic accuracy (AUC = 0.99 vs AUC = 0.86 for CA19-9 alone).
Conclusions: In this study, an exosome-based liquid biopsy signature for the noninvasive and robust detection of patients with PDAC was developed.